|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FRAG | definition of molecular fragment |
| site | atom sites in molecular fragment |
| zmatrix | geometry description matrix |
| rotate | rotational search parameters |
| transl | translational search parameters |
| triple | triple relationship parameters |
| shift | shift coordinates of following atom sites |
| spin | rotate coordinates of following atom sites about Cartesian axes |
| twist | twist following atom sites about connected atom site |
| PATSEE | Option | Code | Arg | Def | |
| do not list bond distances and angles of fragments |
|
| FRAG | 1 | Use cartesian coordinates on site lines. |
|
||
| or | |||||
| 1-3 | cell lengths in Ĺ a, b, c [a] | ||||
| 4-6 | cell angles in degrees
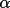 , ,
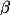 , ,
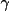 [a]
[a]
|
||||
| or | |||||
| 1- | labels of atoms to be extracted from
lratom:
of the input bdf |
||||
| or | |||||
| 1 | five membered ring |
|
|||
| benzene planar conformation |
|
||||
| benzene boat conformation |
|
||||
| benzene chair conformation |
|
||||
[a] These are applied to the fractional coordinates of the atom sites that follow and must be entered if the cell differs from that on the bdf. | |||||
| site | 1 | atom label | |
| 2-4 | coordinates [a]x, y, z | ||
| 5 | relative site weight in search ( max 1, min 0 ) |
1
|
|
[a] Assumed fractional unless the preceding frag line contains the Cartesian code. | |||
| zmatrix | 1 | n1 sequence number | |
| 2 | atom label | ||
| 3 | n2 sequence number | ||
| 4 | distance to atom n1 | ||
| 5 | n3 sequence number | ||
| 6 | n3-n2-n1 angle | ||
| 5 | n4 sequence number | ||
| 6 | n1-n2-n3 n2-n3-n4 torsion angle |
| rotate | Option | Code | Arg | Def | |
| number of rotations applied |
|
|
auto | ||
| number of solutions saved |
|
|
5
|
||
| intra-mol. vector angle resolution |
|
|
10° |
||
| intra-mol. vector d min/max |
|
|
2.
6.A
|
||
| intra-mol. vectors for Rfom (ratio) |
|
|
0.5
|
||
| intra-mol. vectors for pretest (ratio) |
|
|
0.1
|
||
| Rfom acceptance threshold |
|
|
0.4
|
||
| number of refinement searches |
|
|
1000
|
||
| random rotation start value |
|
|
5431
|
||
| rotation start positions about a b c |
|
|
0°
0 °
0 ° |
||
| rotation range about a b c |
|
|
auto | ||
| list all intra-molecular vectors |
|
no | |||
| signal mirror in fragment |
|
no |
| transl | Option | Code | Arg | Def | |
| number of translations applied |
|
|
auto | ||
| number of solutions saved |
|
|
5
|
||
| inter-mol. vector d minimum |
|
|
3.4
A
|
||
| inter-mol. vector shift resolution |
|
|
0.25
A
|
||
| Tfom acceptance threshold |
|
|
0.3
|
||
| t3sum acceptance threshold |
|
|
0.3
|
||
| translation origin as x/a y/b z/c [a] |
|
|
0 0
0
|
||
| translation range as x/a y/b z/c [a] |
|
|
auto | ||
| list triplets used in t3sum |
|
no | |||
| exclude use of t3sum refinement |
|
include | |||
| random translation start value |
|
|
5431
|
||
[a] Six values may be entered; three for the rotation/translation fragment and optionally three for the translation-only fragment. | |||||
| triple | Option | Code | Arg | Def | |
| minimum E value used |
|
|
auto | ||
| minimum A of any triple |
|
|
auto | ||
| max number of E's used |
|
|
100
|
||
| max number of triples used |
|
|
100
|
| shift [a] | 1-9 | rotation matrix elements
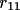 , ,
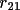 , ,
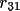 , …, , …,
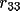
|
|
| 10-12 | translation vector
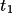 , ,
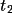 , ,
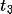
|
||
[a] The shift transformation matrix is applied to all atom sites in the next input fragment. | |||
| spin [a] | 1-3 | rotation angles (in degrees) about the Cartesian axes x, y, z | |
[a] A positive value is an anticlockwise rotation as the axis comes towards you. The spin angles are applied to all atom sites in the next input fragment. | |||
| twist | Option | Code | Arg | Def | |
| minimum torsion angle (deg.) |
|
|
0
|
||
| maximum torsion angle (deg.) |
|
|
355
|
||
| increment torsion angle (deg.) |
|
|
5
|
||
| torsion angle test method |
|
|
1
|