|
| Instructions | Function |
| 101-102 | print distances, angles, and thermal eigenvectors |
| 103 | print thermal eigenvectors and direction cosines |
| 105-106 | same as 101-102 but with convoluting sphere of enclosure |
| 201-202 | define start plot, advance plot, end plot |
| 301-303 | specify plot dimensions, title rotation, retrace |
| 401-407 | define atoms to loaded into the working list for later plotting |
| 411-417 | define atoms to deleted from the working list |
| 501-503 | define plot orientation: axes, rotations, projection axis translation |
| 511 | implement overlap (hidden line) correction |
| 601-604 | define plot position and scaling: set origin, set scale |
| 611-614 | define plot incremental position and scaling |
| 701-715 | specify thermal ellipsoid parameters, draw atom symbols. |
| 801-813 | bond plotting parameters |
| 901-916 | plot labels and titles |
Saved sequences
In some situations, for example plotting a stereoscopic pair, it is necessary to repeat a long series of instructions. By using the saved sequence feature, the long series of instructions needs to be input once only. To store the saved sequence use the following lines:
svstar seq1 : start of saved sequence called seq1 inst ..... : the instructions which make up the inst ..... : saved sequence seq1 svend : end of the saved sequence seq1
To invoke the saved sequence use a control line with the name of the saved sequence use the following line:
svexec seq1
The saved sequence feature is available only for the manual mode.
ANC - Atom Number Code
An atom number code is the sequence number of an atom in the list of reference atoms.
ANR - Atom Number Run
An atom number run is a series of atoms contained in the list of reference atoms. The ANR is specified by designating the atom number codes of the first and last atoms in the run (i.e. ANC1 - ANC2).
ADC - Atom Designator Code
An atom designator code specifies an atom position. It has three components; the atom number, the translation number, and the symmetry equivalent position number. The translation number is a three digit number, where each digit corresponds to a lattice translation along each cell direction with respect to the origin as 555.
ADR - Atom Designator Run
An atom designator run is specified by two atom designator codes. Let ADC1 have components A1, T1, S1 and ADC2 have components A2, T2, S2. The ADR then consists of all atom positions which have A1<=atom number<=A2, T1<=translation number<=T2, and S1<=symmetry number<=S2.
VSC - Vector Search Codes
Vector search codes consist mainly of two atom number runs (origin ANR and target ANR) and a distance range. VSC's place constraints on the search for interatomic vectors. In contrast to previous versions of ORTEP all VSC's are input before any instructions are read in this version. The instructions which use VSC's are 101, 102, 402, 405-407, 412, 415, 416, 801-803, and 811-813.
Height-colouring
It is possible to use the height of each atom above
or below the plane of the drawing to determine its colour.
This height may be found in the atom listing produced by "
genins
" as the z Cartesian coordinate in inches.
The limits for the height classification are introduced on
a "
height " line. An atom
will be assigned to a colour of 1 plus the index of the
largest height limit smaller than the z coordinate of the
atom. Thus if the height limits are arranged in ascending
order, and an atom has a z coordinate larger than limit M
but smaller than limit M+1, the atom will be assigned
colour M+1. If N limits are given (max 8), limits 0 and N+1
always have values of -Infinity and +Infinity
respectively.list
-
Reads
lrcell:and symmetry data from the archive file -
Writes a plot command file on
ort -
Writes a PS300 plot file to
or3
ORTEP mole acta
This is an Acta C style ellipsoid plot of a single molecule. Overlap will be applied. Hydrogens will be treated as spheres of fixed radius. There will be no perspective.
ORTEP cell pers
As above, except that the contents of the unit cell will be generated with a perspective view.
COMPID ICEANE ORTEP molecule sphere over :define type of plot genins cbla list :calculate bond lengths plotp 11 11 24 1 ab *7 70 20 :plot dimensions and axes ellips 6 1.00 :type of ellipsoid to be plotted
Drawing of the molecule iceane (Hamon, et al., 1977).
One iceane molecule is to be plotted. Bond lengths and
angles are to be calculated up to a distance of 2.0
Angstroms. A full listing of atom and bond search output is
to be printed. Plot dimensions are 11 by 11 inches with a 1
inch margin. The view distance is 24 inches. The A and B
axes are to be taken as the X and Y axes, respectively, of
the plot plane. The plot is to be rotated
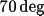 about the X axis
and
about the X axis
and
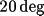 about the Y axis.
The ellipsoids are to be type 6 (with octant shading) with
probability scale 1.00 (i.e. probability that the ellipsoid
encloses the atom is 20 percent).
about the Y axis.
The ellipsoids are to be type 6 (with octant shading) with
probability scale 1.00 (i.e. probability that the ellipsoid
encloses the atom is 20 percent).
title AKERMANNITE - 1.85 ORTEP manu inpu radius or .01 vsc 1 1 1 1 1 .10 1.00 .005 vsc 1 1 2 6 1 .10 2.00 .03 vsc 3 3 1 6 3 .10 1.85 .03 symbol svstar ak inst 511 0 inst 702 inst 801 1 555 1 1 565 1 1 555 1 1 556 1 1 555 1 1 655 1 inst 801 1 666 1 1 665 1 1 666 1 1 656 1 1 666 1 1 566 1 inst 801 1 655 1 1 656 1 1 655 1 1 665 1 1 656 1 1 556 1 inst 801 1 565 1 1 665 1 1 565 1 1 566 1 1 556 1 1 566 1 inst 802 inst 902 *8 0 0 .25 3 -5 .5 1 svend inst 201 inst 301 12 12 30 .5 inst 401 1 555 1 -1 666 1 inst 401 1 555 3 -1 556 3 inst 401 2 555 1 -2 565 1 inst 401 2 565 2 -2 665 2 inst 401 2 655 2 inst 401 2 546 3 -2 556 3 inst 401 2 456 4 -2 556 4 inst 401 2 566 4 inst 401 3 554 1 -3 655 1 inst 401 3 564 2 -3 665 2 inst 401 3 546 3 -3 557 3 inst 401 3 556 4 -3 567 4 inst 402 1 555 1 1 666 1 4 6 2.0 inst 402 1 555 3 1 556 3 4 6 2.0 inst 402 3 554 1 3 655 1 4 6 1.85 inst 402 3 564 2 3 665 2 4 6 1.85 inst 402 3 546 3 3 557 3 4 6 1.85 inst 402 3 556 4 3 567 4 4 6 1.85 inst 501 7 555 1 1 555 1 1 565 1 1 555 1 1 655 1 1 inst 502 1 -75 2 5 inst 601 6 6 .85 1.54 inst 503 2 2.7 svexec ak inst 202 15 inst 503 2 -2.7 svexec ak inst 203
This is an example of an essentially manual run. The
vsc lines set the bonds
to be drawn in the 800 series instruction. Notice that the
.005 in the first
vsc line is a value which
causes a narrow line to be plotted for the lines connecting
the atoms given in the 801 instructions. In this case, this
line outlines the unit cell. In the
symbol line, the name is
placed to the right to cause it to be centred on the plot.
svstar marks the start of
the instructions that are to be saved for use. This
placement of the 511 through 902 instructions is distinctly
different from that order used for Johnson's version of
ORTEP. The
svend line marks the end
of the saved instructions. The next
inst lines are executed
as encountered. The
svexec line causes the
saved (name
) instructions to be carried out. The reason
they are done twice is to produce the stereo pair. If there
was no stereo plot, the 511-902 would not need to be saved
and could have been placed in order.ak
-
Johnson, C.K., Guerdon, J.F., Richard, P., Whitlow, S., and Hall, S.R. 1972. ORTEP. The X-RAY system of Crystallographic Programs. TR-192, 283p.
-
Johnson, C.K. 1970. ORTEP: A FORTRAN Thermal-Ellipsoid Plot Program for Crystal Structure Illustrations., Report ORNL-3794, 135 pages.
-
Johnson, C.K. 1971. Supplementary Instructions for the Oak Ridge Thermal Ellipsoid Plot Program ORTEP-II., Oak Ridge National Laboratory., ORNL-3794 supp., 10 pages.